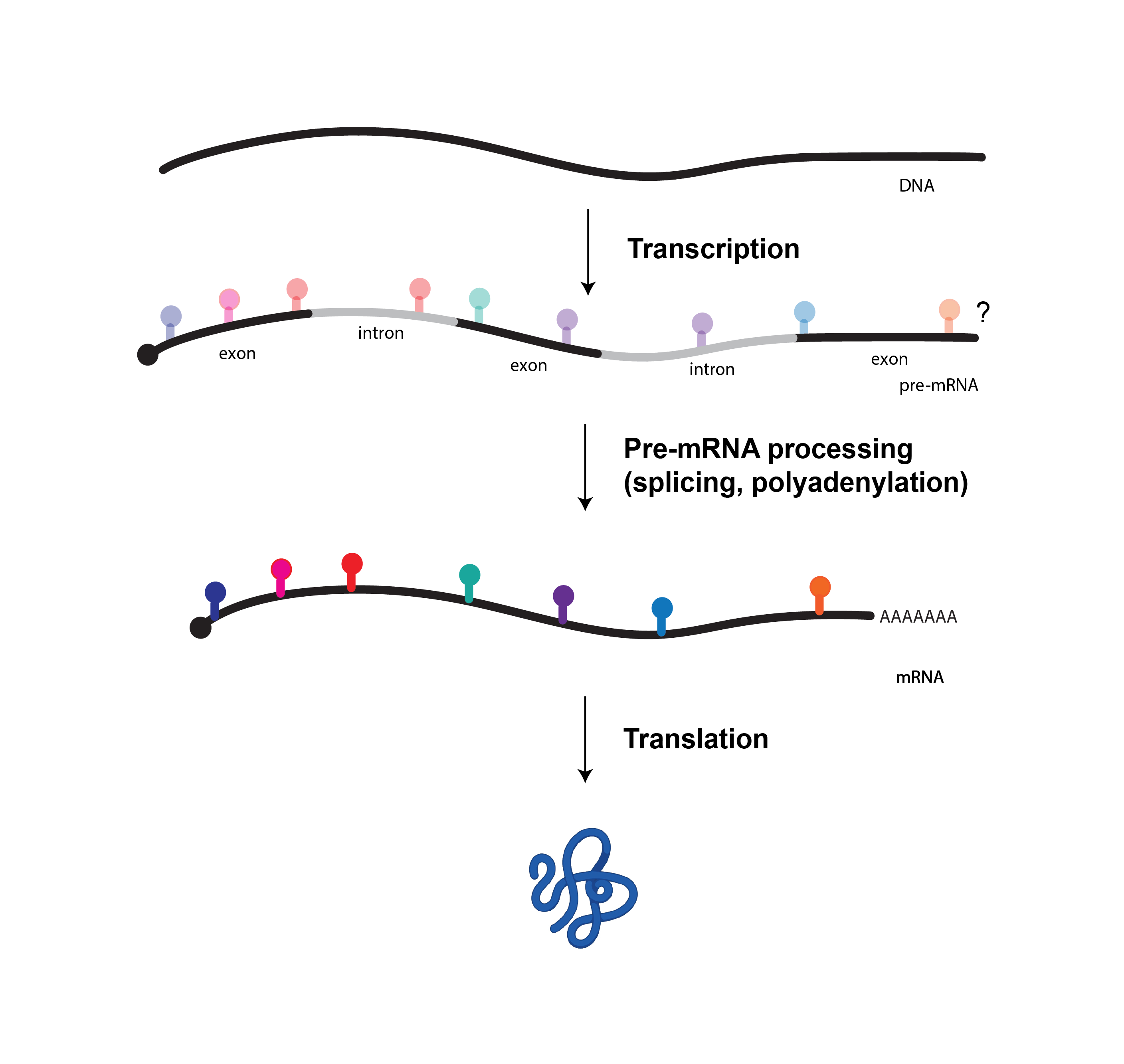
Our lab is broadly interested in RNA-based mechanisms of gene regulation. Precise control of gene expression at the level of messenger RNA processing is necessary for organismal development, required for response to environmental cues and its dysregulation is the basis of many diseases. We are keen to uncover mechanisms that control alternative mRNA processing and their downstream consequences on gene expression and cell physiology. Eukaryotic mRNA are extensively modified with non-canonical bases that have the potential to regulate pre-mRNA processing steps such as splicing and 3’ end processing. Dysregulation of RNA-modifying enzymes cause a wide range of human diseases, underscoring the need to elucidate this exciting new layer of gene regulation. Our current research studies mechanisms and functions of RNA modifications in pre-mRNA processing and their roles in development and disease through a combination of molecular biology, biochemistry, genomics, genetics, and systems biology.
We uncovered that pseudouridine is added to nascent pre-mRNA co-transcriptionally by multiple pseudouridine synthases, including several that are implicated in human disease. This finding demonstrates that pseudouridine is in place to influence all steps of mRNA metabolism including nuclear processing. Our work revealed a novel function for pseudouridine synthases in alternative splicing regulation and pre-mRNA 3′ end processing events in human cells and demonstrated that individual pseudouridines can directly impact splicing. We are interested in defining the underlying mechanisms and physiological functions of co-transcriptional pre-mRNA modifications. Our initial focus is on pseudouridine, which will serve as a prototype for investigating the function of other RNA modifications and will inform our understanding of the many diseases caused by pseudouridine synthases.
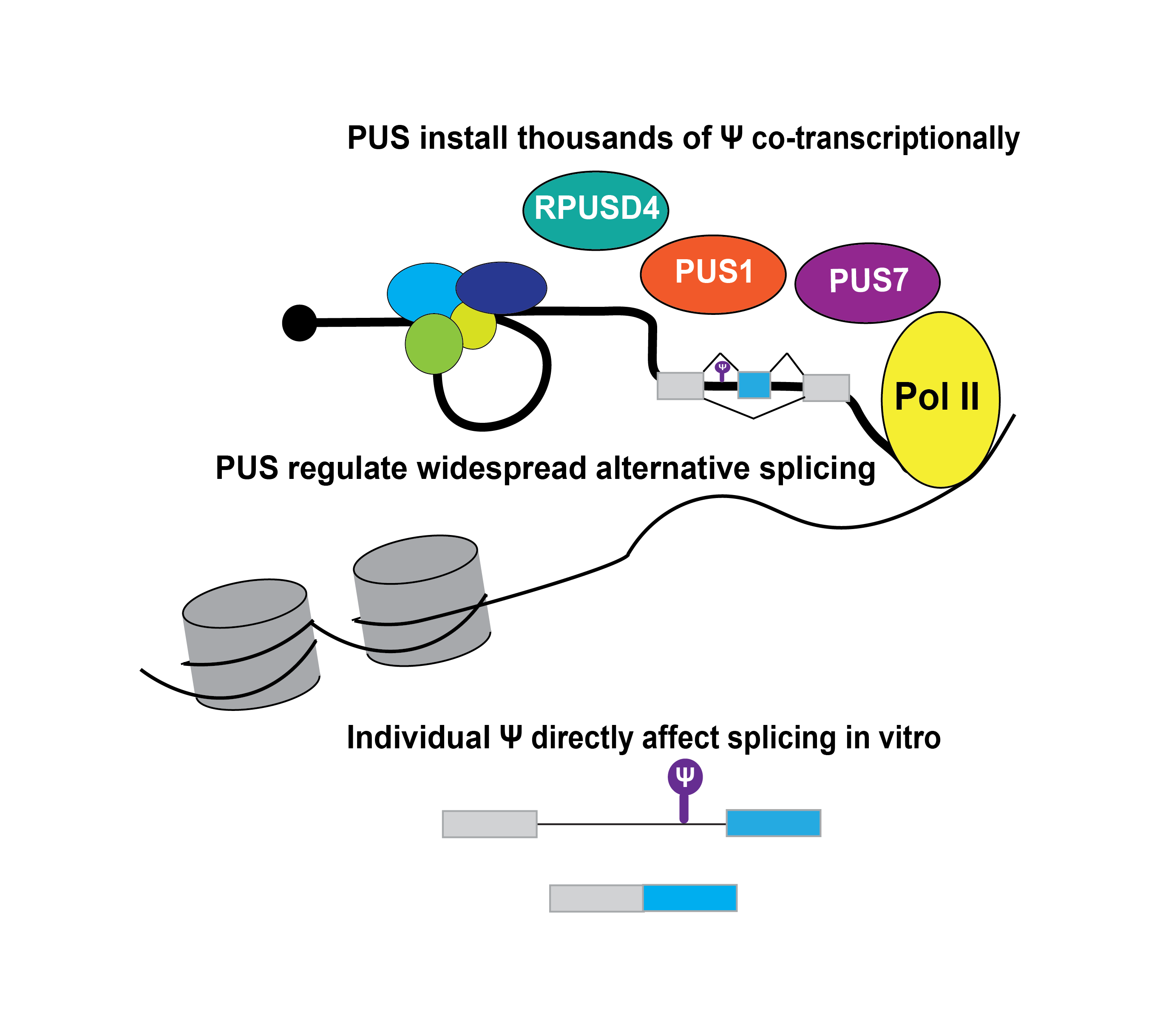
Pseudouridine deposition is regulated in a cell type-specific manner and in response to cellular stresses. It is unclear why certain pre-mRNA regions are targeted for modification with pseudouridine and not others. Work in our lab is investigating the determinants of co-transcriptional pseudouridylation.
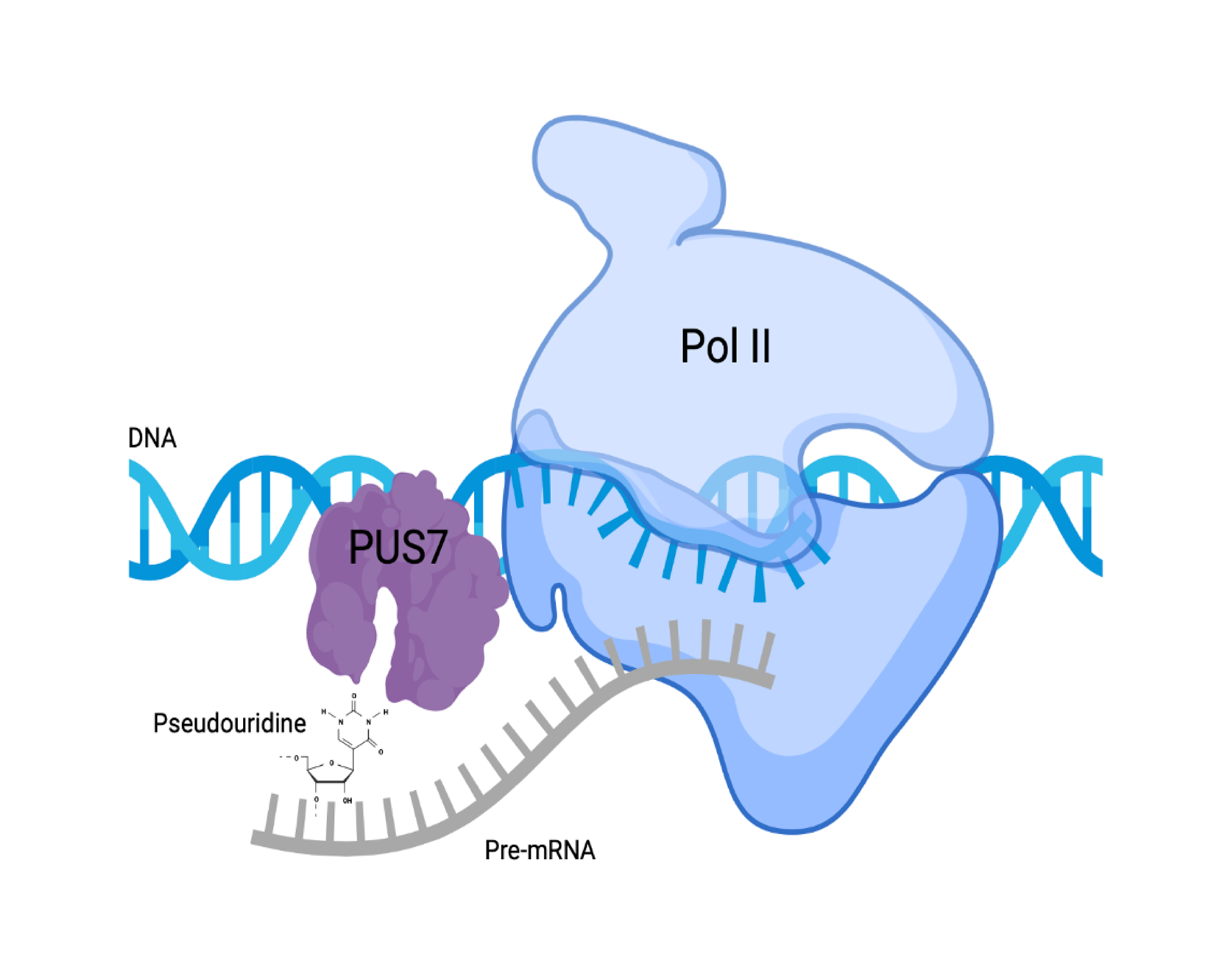
Figure created with BioRender.
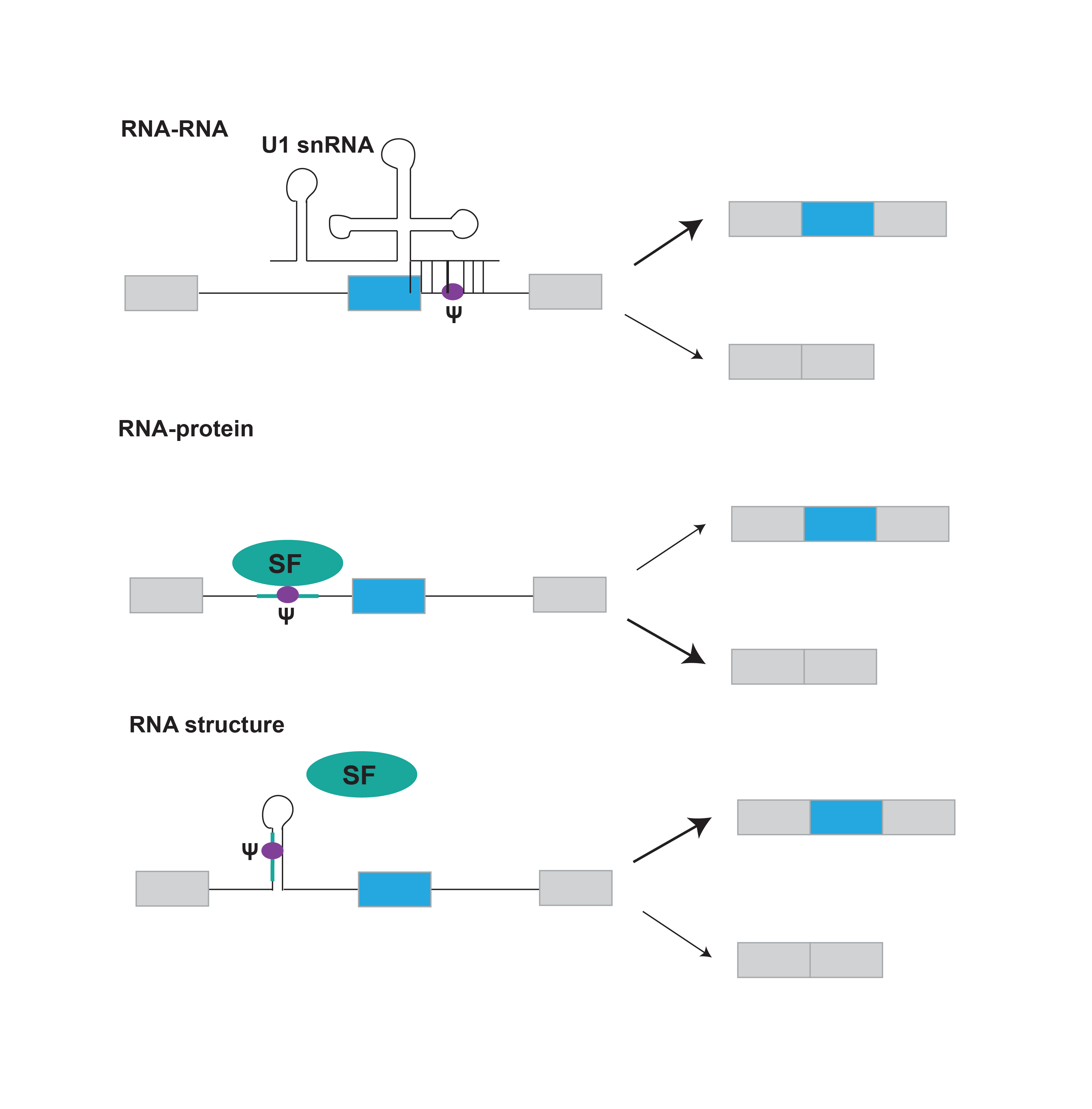
Pseudouridines in pre-mRNA have the potential to influence splicing by three main mechanisms: directly by modulating pre-mRNA-protein interactions, indirectly by influencing pre-mRNA secondary structure to modulate accessibility of regulatory sequences, or by altering pre-mRNA-spliceosomal snRNA interactions. We will elucidate by which of these mechanisms individual pseudouridines (Ψ ) in pre-mRNA affect splicing and other pre-mRNA processing steps.
RNA modifying enzymes are known to cause or are associated with a wide range of human diseases, including developmental disorders and cancer. One of our goals is to connect molecular functions of RNA modifications to cellular and organismal phenotypes. Future work will investigate the function of RNA modifying enzymes and disease-causing variants in cellular differentiation and development.
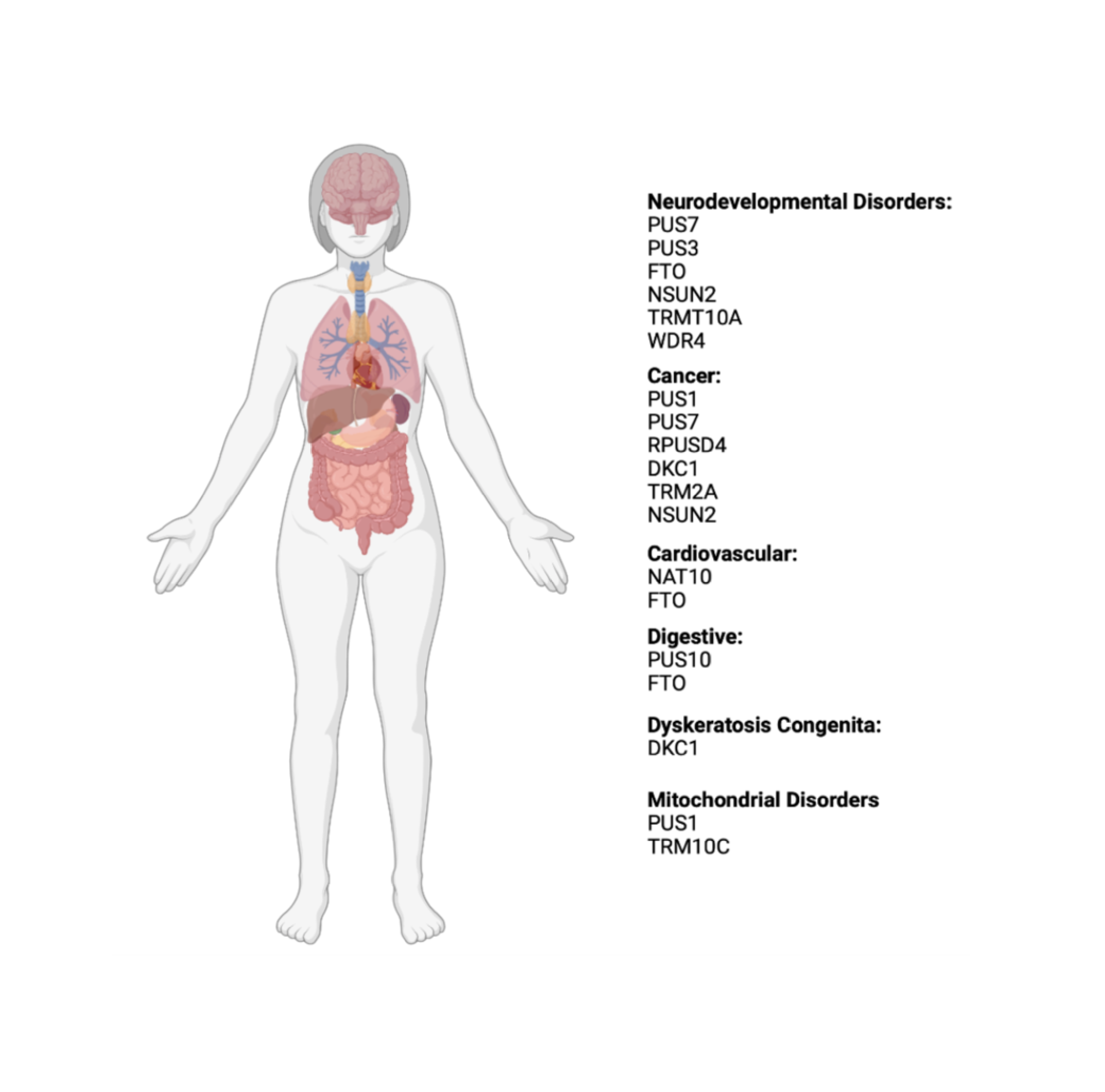
Figure created with BioRender.
New highthroughput methods have revealed widespread decoration of eukaryotic messenger RNA with diverse RNA modifications whose functions in mRNA metabolism and gene expression are largely unknown. We are interested in defining the broader landscape of RNA modifications in pre-mRNA by using/developing cutting edge tools for modification detection and functional characterization.